Home Web>SubgenomeEvolution (06 Nov 2025)
How subgenomes evolve
Parental components in hybrid plant genomes
The Institute of Computational Biology developed a method to measure the intermixing state of subgenomes in polyploid plants. The work published in The Plant Journal (Schiavinato et al. 2021) testifies to the power of bioinformatics for obtaining insights into important biological processes solely by applying novel data analysis strategies. Interestingly, the taxonomic age of a hybrid does not seem to correlate with the degree of subgenome conservation. The genetic information collectively referred to as "the genome" of an extant organism may be the product of a very complex history. It is well known by now that many species were generated by ancient hybridisation events. By crossing of two separate species which have remained interbreedable despite their status as different taxa, novel species can be generated. This pathway for species generation is particularly common for plants. Examples of such hybrid species include rapeseed, tobacco, cotton, quinoa and many more. Intriguingly, these hybrid plant species encompass the genetic information of both of their parents and initially carry four rather than just two sets of chromosomes. The study started with the authors' previous observation that the parental chromosomes in one such ancient hybrid, a tobacco species native to Australia, were poorly retained in their original condition. Yet, in quinoa, another hybrid species of similar age the level of conservation was quite high and the assignment of individual quinoa chromosomes to their presumptive parents was straightforward. Thus, the forces that may act onto the genome of a hybrid species can differ remarkably between taxa, resulting in different degrees of intermixing of the parental chromosomes. In the current study, the authors used publicly available sequencing data to shed light onto how such intermixing compares to hybrid age, to its domestication status, to its reproductive strategy, and to particular genome features. The authors believe that their comprehensive analysis of ten economically relevant plant species could pioneer larger scale studies to further establish correlations between genome stability in hybrids and other biological processes.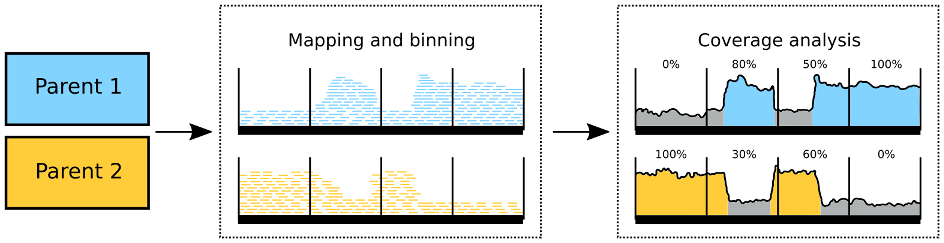
Publication
Schiavinato* M, Bodrug-Schepers* A, Dohm JC, Himmelbauer H (2021). Subgenome evolution in allotetraploid plants. The Plant Journal (2021) 106, 672–688. https://doi.org/10.1111/tpj.15190. *shared first authorshipSoftware
https://github.com/MatteoSchiavinato/manticore.git| 07 Mar 2026 - 11:06 | Foswiki v2.0.2 |
